Random Trees¶
Random trees have been introduced by Leo Breiman and Adele Cutler: http://www.stat.berkeley.edu/users/breiman/RandomForests/ . The algorithm can deal with both classification and regression problems. Random trees is a collection (ensemble) of tree predictors that is called forest further in this section (the term has been also introduced by L. Breiman). The classification works as follows: the random trees classifier takes the input feature vector, classifies it with every tree in the forest, and outputs the class label that recieved the majority of “votes”. In the case of regression the classifier response is the average of the responses over all the trees in the forest.
All the trees are trained with the same parameters, but on the different training sets, which are generated from the original training set using the bootstrap procedure: for each training set we randomly select the same number of vectors as in the original set (
=N
). The vectors are chosen with replacement. That is, some vectors will occur more than once and some will be absent. At each node of each tree trained not all the variables are used to find the best split, rather than a random subset of them. With each node a new subset is generated, however its size is fixed for all the nodes and all the trees. It is a training parameter, set to
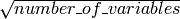 by default. None of the trees that are built are pruned.
by default. None of the trees that are built are pruned.
In random trees there is no need for any accuracy estimation procedures, such as cross-validation or bootstrap, or a separate test set to get an estimate of the training error. The error is estimated internally during the training. When the training set for the current tree is drawn by sampling with replacement, some vectors are left out (so-called oob (out-of-bag) data ). The size of oob data is about N/3 . The classification error is estimated by using this oob-data as following:
- Get a prediction for each vector, which is oob relatively to the i-th tree, using the very i-th tree.
- After all the trees have been trained, for each vector that has ever been oob, find the class-“winner” for it (i.e. the class that has got the majority of votes in the trees, where the vector was oob) and compare it to the ground-truth response.
- Then the classification error estimate is computed as ratio of number of misclassified oob vectors to all the vectors in the original data. In the case of regression the oob-error is computed as the squared error for oob vectors difference divided by the total number of vectors.
References:
Machine Learning, Wald I, July 2002.
Looking Inside the Black Box, Wald II, July 2002.
Software for the Masses, Wald III, July 2002.
And other articles from the web site http://www.stat.berkeley.edu/users/breiman/RandomForests/cc_home.htm .
CvRTParams¶
- CvRTParams¶
Training Parameters of Random Trees.
struct CvRTParams : public CvDTreeParams
{
bool calc_var_importance;
int nactive_vars;
CvTermCriteria term_crit;
CvRTParams() : CvDTreeParams( 5, 10, 0, false, 10, 0, false, false, 0 ),
calc_var_importance(false), nactive_vars(0)
{
term_crit = cvTermCriteria( CV_TERMCRIT_ITER+CV_TERMCRIT_EPS, 50, 0.1 );
}
CvRTParams( int _max_depth, int _min_sample_count,
float _regression_accuracy, bool _use_surrogates,
int _max_categories, const float* _priors,
bool _calc_var_importance,
int _nactive_vars, int max_tree_count,
float forest_accuracy, int termcrit_type );
};
The set of training parameters for the forest is the superset of the training parameters for a single tree. However, Random trees do not need all the functionality/features of decision trees, most noticeably, the trees are not pruned, so the cross-validation parameters are not used.
CvRTrees¶
- CvRTrees¶
Random Trees.
class CvRTrees : public CvStatModel
{
public:
CvRTrees();
virtual ~CvRTrees();
virtual bool train( const CvMat* _train_data, int _tflag,
const CvMat* _responses, const CvMat* _var_idx=0,
const CvMat* _sample_idx=0, const CvMat* _var_type=0,
const CvMat* _missing_mask=0,
CvRTParams params=CvRTParams() );
virtual float predict( const CvMat* sample, const CvMat* missing = 0 )
const;
virtual void clear();
virtual const CvMat* get_var_importance();
virtual float get_proximity( const CvMat* sample_1, const CvMat* sample_2 )
const;
virtual void read( CvFileStorage* fs, CvFileNode* node );
virtual void write( CvFileStorage* fs, const char* name );
CvMat* get_active_var_mask();
CvRNG* get_rng();
int get_tree_count() const;
CvForestTree* get_tree(int i) const;
protected:
bool grow_forest( const CvTermCriteria term_crit );
// array of the trees of the forest
CvForestTree** trees;
CvDTreeTrainData* data;
int ntrees;
int nclasses;
...
};
CvRTrees::train¶
- bool CvRTrees::train(const CvMat* train_data, int tflag, const CvMat* responses, const CvMat* comp_idx=0, const CvMat* sample_idx=0, const CvMat* var_type=0, const CvMat* missing_mask=0, CvRTParams params=CvRTParams())¶
- Trains the Random Trees model.
The method CvRTrees::train is very similar to the first form of CvDTree::train () and follows the generic method CvStatModel::train conventions. All of the specific to the algorithm training parameters are passed as a CvRTParams instance. The estimate of the training error ( oob-error ) is stored in the protected class member oob_error .
CvRTrees::predict¶
- double CvRTrees::predict(const CvMat* sample, const CvMat* missing=0) const¶
- Predicts the output for the input sample.
The input parameters of the prediction method are the same as in CvDTree::predict , but the return value type is different. This method returns the cumulative result from all the trees in the forest (the class that receives the majority of voices, or the mean of the regression function estimates).
CvRTrees::get_var_importance¶
The method returns the variable importance vector, computed at the training stage when :ref:`CvRTParams`::calc_var_importance is set. If the training flag is not set, then the NULL pointer is returned. This is unlike decision trees, where variable importance can be computed anytime after the training.
CvRTrees::get_proximity¶
- float CvRTrees::get_proximity(const CvMat* sample_1, const CvMat* sample_2) const¶
- Retrieves the proximity measure between two training samples.
The method returns proximity measure between any two samples (the ratio of the those trees in the ensemble, in which the samples fall into the same leaf node, to the total number of the trees).
Example: Prediction of mushroom goodness using random trees classifier
#include <float.h>
#include <stdio.h>
#include <ctype.h>
#include "ml.h"
int main( void )
{
CvStatModel* cls = NULL;
CvFileStorage* storage = cvOpenFileStorage( "Mushroom.xml",
NULL,CV_STORAGE_READ );
CvMat* data = (CvMat*)cvReadByName(storage, NULL, "sample", 0 );
CvMat train_data, test_data;
CvMat response;
CvMat* missed = NULL;
CvMat* comp_idx = NULL;
CvMat* sample_idx = NULL;
CvMat* type_mask = NULL;
int resp_col = 0;
int i,j;
CvRTreesParams params;
CvTreeClassifierTrainParams cart_params;
const int ntrain_samples = 1000;
const int ntest_samples = 1000;
const int nvars = 23;
if(data == NULL || data->cols != nvars)
{
puts("Error in source data");
return -1;
}
cvGetSubRect( data, &train_data, cvRect(0, 0, nvars, ntrain_samples) );
cvGetSubRect( data, &test_data, cvRect(0, ntrain_samples, nvars,
ntrain_samples + ntest_samples) );
resp_col = 0;
cvGetCol( &train_data, &response, resp_col);
/* create missed variable matrix */
missed = cvCreateMat(train_data.rows, train_data.cols, CV_8UC1);
for( i = 0; i < train_data.rows; i++ )
for( j = 0; j < train_data.cols; j++ )
CV_MAT_ELEM(*missed,uchar,i,j)
= (uchar)(CV_MAT_ELEM(train_data,float,i,j) < 0);
/* create comp_idx vector */
comp_idx = cvCreateMat(1, train_data.cols-1, CV_32SC1);
for( i = 0; i < train_data.cols; i++ )
{
if(i<resp_col)CV_MAT_ELEM(*comp_idx,int,0,i) = i;
if(i>resp_col)CV_MAT_ELEM(*comp_idx,int,0,i-1) = i;
}
/* create sample_idx vector */
sample_idx = cvCreateMat(1, train_data.rows, CV_32SC1);
for( j = i = 0; i < train_data.rows; i++ )
{
if(CV_MAT_ELEM(response,float,i,0) < 0) continue;
CV_MAT_ELEM(*sample_idx,int,0,j) = i;
j++;
}
sample_idx->cols = j;
/* create type mask */
type_mask = cvCreateMat(1, train_data.cols+1, CV_8UC1);
cvSet( type_mask, cvRealScalar(CV_VAR_CATEGORICAL), 0);
// initialize training parameters
cvSetDefaultParamTreeClassifier((CvStatModelParams*)&cart_params);
cart_params.wrong_feature_as_unknown = 1;
params.tree_params = &cart_params;
params.term_crit.max_iter = 50;
params.term_crit.epsilon = 0.1;
params.term_crit.type = CV_TERMCRIT_ITER|CV_TERMCRIT_EPS;
puts("Random forest results");
cls = cvCreateRTreesClassifier( &train_data,
CV_ROW_SAMPLE,
&response,
(CvStatModelParams*)&
params,
comp_idx,
sample_idx,
type_mask,
missed );
if( cls )
{
CvMat sample = cvMat( 1, nvars, CV_32FC1, test_data.data.fl );
CvMat test_resp;
int wrong = 0, total = 0;
cvGetCol( &test_data, &test_resp, resp_col);
for( i = 0; i < ntest_samples; i++, sample.data.fl += nvars )
{
if( CV_MAT_ELEM(test_resp,float,i,0) >= 0 )
{
float resp = cls->predict( cls, &sample, NULL );
wrong += (fabs(resp-response.data.fl[i]) > 1e-3 ) ? 1 : 0;
total++;
}
}
printf( "Test set error =
}
else
puts("Error forest creation");
cvReleaseMat(&missed);
cvReleaseMat(&sample_idx);
cvReleaseMat(&comp_idx);
cvReleaseMat(&type_mask);
cvReleaseMat(&data);
cvReleaseStatModel(&cls);
cvReleaseFileStorage(&storage);
return 0;
}
Help and Feedback
You did not find what you were looking for?- Try the FAQ.
- Ask a question in the user group/mailing list.
- If you think something is missing or wrong in the documentation, please file a bug report.
